Our two main areas of research are: ER bodies as chemical defence system in plants and autophagy and quality control of peroxisomes.
- ER bodies as chemical defence system in plants
- Autophagy and quality control of peroxisomes
Sessile plants have no choice in the environment where they are growing. Therefore, plants have developed sophisticated defence systems to adapt to changes happening in the environment. We discovered ER bodies and their involvement in the so-called “mustard oil bomb” defence system against animal herbivores. The system works by accumulating β-glucosidase (BGLU), which is necessary to activate defensive glucosinolates (Figure 1).
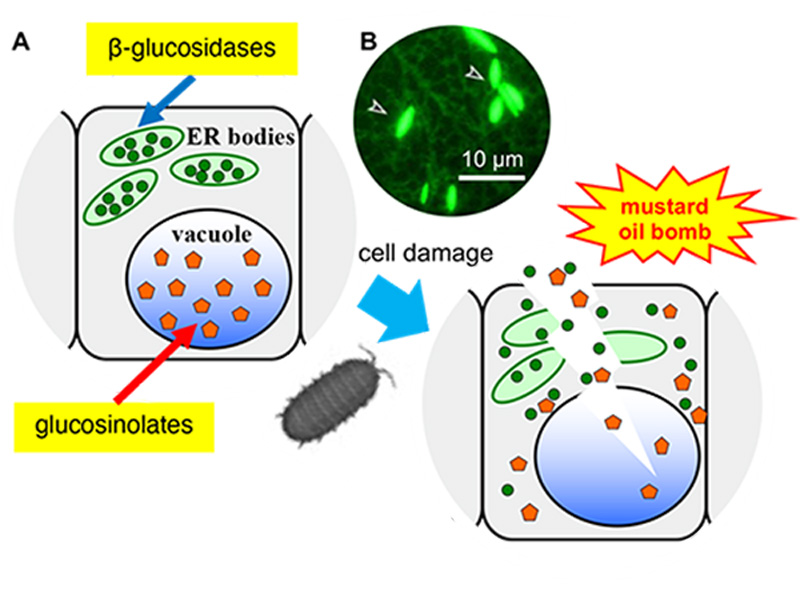
Figure 1. Arabidopsis thaliana has a defence system based on ER bodies.
(A) ER bodies accumulate β-glucosidase (green circle), and vacuoles accumulate the substrates, glucosinolates (orange pentagon). The enzyme and substrate come to contact and produce toxic molecules immediately after the feeding damage.
(B) Fluorescent image of transgenic plants expressing ER-targeted GFP. Arrowheads indicate ER bodies.
ER bodies are observed in plants of the order Brassicales, including a model plant Arabidopsis thaliana. A transgenic Arabidopsis expressing green fluorescent protein (GFP) in the endoplasmic reticulum (ER), spindle-shaped ER bodies were observed in addition to the regular ER network (Figure 1). Further analysis of the ER body defence system revealed that the ER bodies could be divided into two types. Constitutive-type ER bodies (cER bodies) accumulate in seedlings and roots, and inducible-type ER bodies (iER bodies) are induced accumulation after wounding in rosette leaves through the action of methyl jasmonate (a wounding hormone in plants). Brassicaceae plants seem to have evolved various defence systems that increase the defence level and enable the flexible adjustment to different levels of threat.
Gene regulatory system for ER body formation has been deeply analysed (Figure 2). A basic helix-loop-helix (bHLH) type transcription factor bHLH020/NAI1 regulates cER body formation in seedlings and roots. bHLH020/NAI1 regulates the expression of genes encoding ER body proteins, such as those encoded by BGLU23/PYK10, NAI2, MEMBRANE OF ER BODY (MEB) 1 and MEB2 in the seedlings and roots. BGLU23/PYK10 is able to hydrolyse glucosinolates to produce repellent end products, such as isothiocyanates. An ER body protein, NAI2 is necessary for the cER body formation in seedlings and roots, and NAI2 homologues are only observed in the plants of order Brassicales that produce ER bodies. MEB1 and MEB2 have multiple membrane-spanning regions and accumulate on ER body membranes.
A NAI2 homologue, TSA1 is involved in the iER body formation in Arabidopsis rosette leaves. It is shown that iER bodies accumulate BGLU18. TSA1 and BGLU18 expression is induced by the methyl jasmonate treatment with a MYC2/3/4 dependent manner. It is suggested that At-α whole-genome duplication generate NAI2 and TSA1, and therefore two types of ER body formation (cER and iER bodies) mechanism are developed in the Brassicaceae plants.
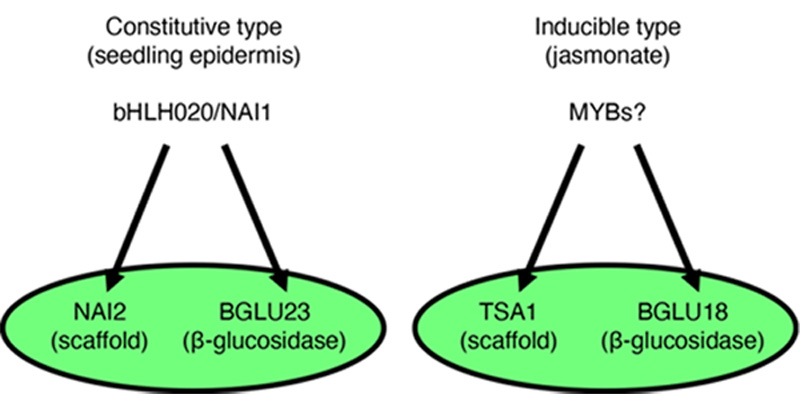
Figure 2. Two types of ER body formation mechanism.
bHLH020/NAI1 regulates NAI2 and BGLU23/PYK10 for the cER body formation in seedlings and roots, while TSA1 and BGLU18 expressed in jasmonate treated leaves to form iER bodies.
MEB1 and MEB2 have a homology to VACUOLAR IRON TRANSPORTER 1 (VIT1), and they have an iron and manganese transport activity. We developed an image analysis tool for microscope images of ER bodies and found that meb1 meb2 mutant showed rounder and less movement of ER bodies. The finding suggested ER body membrane proteins are involved in ER body morphology and dynamics.
Nakano RT, Yamada K, Bednarek P, Nishimura M, Hara-Nishimura I (2014). ER bodies in plants of the Brassicales order: Biogenesis and association with innate immunity. Front. Plant Sci. 5, 73.
Yamada K, Hara-Nishimura I, Nishimura M. (2011) Unique defense strategy by the endoplasmic reticulum body in plants. Plant Cell Physiol. 52, 2039-2049.
Basak AK, Piasecka A, Hucklenbroich J, Türksoy GM, Guan R, Zhang P, Getzke F, Garrido-Oter R, Hacquard S, Strzałka K, Bednarek P, Yamada K, Nakano RT (2022). Tryptophan specialized metabolism and ER body-resident myrosinases modulate root microbiota assembly. bioRxiv (https://doi.org/10.1101/2022.07.06.498822)
Basak AK, Mirzaei M, Strzałka K, Yamada K (2021). Texture feature extraction from microscope images enables a robust estimation of ER body phenotype in Arabidopsis. Plant Methods 17, 109.
Sarkar S, Stefanik N, Kunieda T, Hara-Nishimura I, Yamada K (2021). The Arabidopsis transcription factor NAI1 activates the NAI2 promoter by binding to the G-box motifs. Plant Signal. Behav. 16, 1846928.
Yamada K, Goto-Yamada S, Nakazaki A, Kunieda T, Kuwata K, Nagano AJ, Nishimura M, Hara-Nishimura I. (2020). Endoplasmic reticulum-derived bodies enable a single-cell chemical defense in Brassicaceae plants. Commun. Biol. 3, 21.
Stefanik N, Bizan J, Wilkens A, Tarnawska-Glatt K, Goto-Yamada S, Strzałka K, Nishimura M, Hara-Nishimura I, Yamada K (2020). NAI2 and TSA1 drive differentiation of constitutive and inducible ER body formation in Brassicaceae. Plant Cell Physiol. 61, 722-734.
Nakazaki A, Yamada K, Kunieda T, Tamura K, Hara-Nishimura I, Shimada T (2019). Biogenesis of leaf endoplasmic reticulum body is regulated by both jasmonate-dependent and independent pathways. Plant Signal. Behav. 14, e1622982.
Nakazaki A, Yamada K, Kunieda T, Sugiyama R, Hirai YM, Tamura K, Hara-Nishimura I, Shimada T (2019). Leaf endoplasmic reticulum bodies identified in Arabidopsis rosette leaves are involved in defense against herbivory. Plant Physiol. 179, 1515-1524.
Nakano RT, Piślewska-Bednarek M, Yamada K, Edger PP, Miyahara M, Kondo M, Böttcher C, Mori M, Nishimura M, Schulze-Lefert P, Hara-Nishimura I, Bednarek P (2017). PYK10 myrosinase reveals a functional coordination between ER bodies and glucosinolates in Arabidopsis thaliana. Plant J. 89, 204-220.
Yamada K, Nagano AJ, Nishina M, Hara-Nishimura I, Nishimura M (2013) Identification of two novel endoplasmic reticulum body-specific integral membrane proteins. Plant Physiol. 161, 108-120.
Ogasawara K, Yamada K, Christeller JT, Kondo M, Hatsugai N, Hara-Nishimura I, Nishimura M (2009). Constitutive and inducible ER bodies of Arabidopsis thaliana accumulate distinct β-glucosidases. Plant Cell Physiol. 50, 480-488
Yamada K, Nagano AJ, Nishina M, Hara-Nishimura I, Nishimura M (2008) NAI2 is an endoplasmic reticulum body component that enables ER body formation in Arabidopsis thaliana. Plant Cell 20, 2529-2540.
Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I (2001) A protease-storing body that prepares for cell death or stress in the epidermal cells of Arabidopsis. Plant Cell Physiol. 42, 894-899.
Mechanisms to remove useless and/or toxic cellular components are found in various organelles and cellular processes during maintenance of homeostasis of organisms. Peroxisomes, one of the ubiquitous organelles found in eukaryotic cells, contain appropriate enzymes and change metabolic systems depending on cellular state, plant developmental stage and environmental stimuli. During this functional transition, useless enzymes are degraded rapidly. In addition, peroxisomes produce hydrogen peroxide (H2O2) in the course of their metabolism. Since H2O2 can be the source of the most highly reactive and toxic form of reactive oxygen species (ROS), peroxisomal proteins must inevitably be damaged. Therefore, a quality control system to remove abnormal and toxic proteins is crucial for the maintenance of optimal performance of peroxisomes.
In a previous study, we demonstrated that there are two important actors in the quality control of plant peroxisomes, LON proteinase 2 (LON2) and autophagy (Figure 3). LON2 is one of the protease inside peroxisomes. The C-terminal peptidase domain of LON2 contributes to degradation of unnecessary peroxisomal proteins during the functional transition of peroxisomes. On the other hand, autophagy degrades peroxisomes whole. We showed that autophagy is responsible for degradation of peroxisomes that are highly oxidized and damaged by H2O2. Interestingly, the N-terminus chaperone domain of LON2 is responsible for suppression of peroxisome degradation by autophagy. The present study aims to understand the role of LON2 in peroxisome maintenance and the molecular mechanism whereby autophagy recognizes damaged peroxisomes using Arabidopsis and liverwort Marchantia polymorpha.
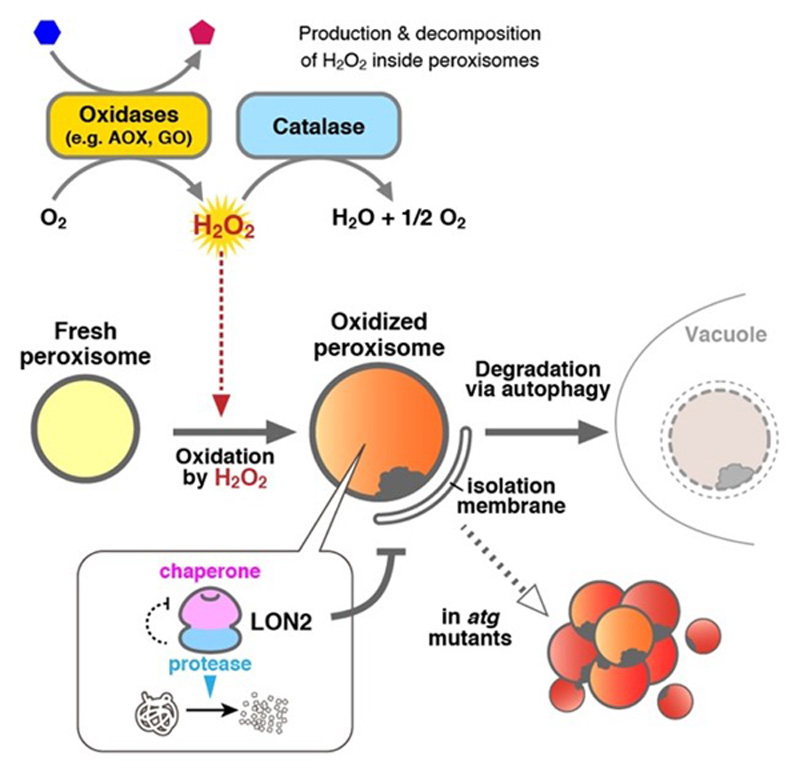
Figure 3. Schematic model of the peroxisome quality control.
Generally, the term autophagy represents "macro-autophagy", in which degradation targets are surrounded by the isolation membrane in the cytosol and transported to the vacuole. Recently we have reported starvation-induced "micro-autophagy" in plant cells, in which the vacuolar membrane directly engulfs and takes degradation targets into the vacuole (Figure 4). Unlike ordinary well-known macroautophagy, little is known about microautophagy. We are studying the molecular mechanism of microautophagy using Arabidopsis and tobacco BY-2 cultured cells.
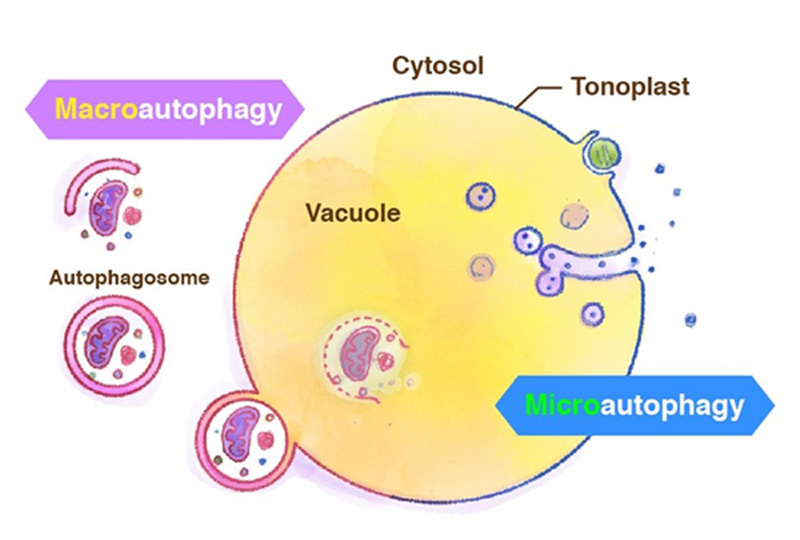
Figure 4. Macroautophagy and microautophagy.
Sieńko K, Poormassalehgoo A, Yamada K, Goto-Yamada S (2020). Microautophagy in plants: consideration of its molecular mechanism. Cells 9, 887.
Goto-Yamada S, Mano S, Yamada K, Oikawa K, Hosokawa Y, Hara-Nishimura I, Nishimura M (2015). Dynamics of the light-dependent transition of plant peroxisomes. Plant Cell Physiol. 56, 1264-1271.
Oikawa K, Goto-Yamada S, Hayashi Y, Takahashi D, Kimori Y, Shibata M, Yoshimoto K, Takemiya A, Kondo M, Hikino K, Kato A, Shimoda K, Ueda H, Uemura M, Numata K, Ohsumi Y, Hara-Nishimura I, Mano S, Yamada K, Nishimura M (2022). Pexophagy suppresses ROS-induced damage in leaf cells under high-intensity light. Nat. Commun. 13, 7493
Goto-Yamada S, Oikawa K, Bizan J, Shigenobu S, Yamaguchi K, Mano S, Hayashi M, Ueda H, Hara-Nishimura I, Nishimura M, Yamada K (2019). Sucrose starvation induces microautophagy in plant root cells. Front. Plant Sci. 10, 1604.
Goto-Yamada S, Mano S, Nakamori C, Kondo M, Yamawaki R, Kato A, Nishimura M. (2014) Chaperone and protease functions of LON protease 2 modulate the peroxisomal transition and degradation with autophagy. Plant Cell Physiol. 55, 482-496.
Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, Hayashi M, Sakamoto W, Ohsumi Y, Nishimura M (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25, 4967-4983.
Goto S, Mano S, Nakamori C, Nishimura M (2011). Arabidopsis ABERRANT PEOXISOME MORPHOLOGY 9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell 23, 1537-1587.

Kenji Yamada, PhD
Read More o Kenji Yamada, PhDKenji Yamada (group leader, 2016-)
 Katarzyna Tarnawska-Glatt (postdoc, 2016-)
Katarzyna Tarnawska-Glatt (postdoc, 2016-)
 Shino Goto-Yamada (postdoc, project PI, 2016-)
Shino Goto-Yamada (postdoc, project PI, 2016-)
Kaichiro Endo (postdoc, project PI; 2018-)
Subhankar Bera (postdoc, 2021-)
Kritika Bhardwaj (postdoc, 2022-)
 Rituraj Batth (postdoc, 2022-)
Rituraj Batth (postdoc, 2022-)
 Natalia Stefanik (PhD student, 2016-)
Natalia Stefanik (PhD student, 2016-)
 Alwine Wilkens (PhD student, project PI, 2018-)
Alwine Wilkens (PhD student, project PI, 2018-)
Arpan Kumar Basak (PhD student, project PI, 2017-)
 Mohamadreza Mirzaei (PhD student, 2018-)
Mohamadreza Mirzaei (PhD student, 2018-)
Andisheh Masalehgou (PhD student, 2018-)
Elżbieta Borlik (PhD student, 2020-)
Karolina Małek (technician, 2018-)

Foto: Zespół Yamada Lab, Kwiecień 2022
Mohamadreza Mirzaei NCN PRELUDIUM 22 (2024-2025). Physiological relationship between vascular tissues and ER body-producing cells in Arabidopsis. UMO-2023/49/N/NZ3/02771
Kaichiro Endo NCN SONATA 17 (2022-2025). Exploring the novel vacuolar transporters of glucosinolates for supporting plant defense system against herbivores and pests in Arabidopsis thaliana. UMO-2021/43/D/NZ3/03222
Arpan Kumar Basak, NCN PRELUDIUM 20 (2022-2025). The other side of ER bodies: Revealing the function of ER body membrane proteins in Arabidopsis thaliana. UMO-2021/41/N/NZ3/04537.
Kenji Yamada, NCN OPUS 19 (2021-2025). Gene regulatory mechanism for the mustard oil bomb defence system of ER bodies in Brassicaceae. DEC-2020/37/B/NZ3/04176.
Alwine Wilkens, NCN PRELUDIUM 19 (2021-2023). On the shoulders of giants - How giant cells influence ER body formation. UMO-2020/37/N/NZ3/03591.
Shino Goto-Yamada, NCN SONATA 15 (2020-2023). Unveiling the molecular mechanism of microautophagy in plants. UMO-2019/35/D/NZ3/04500
Shino Goto-Yamada, NCN SONATA-BIS 9 (2020-2024). Molecular insight into the mechanism of peroxisome degradation via autophagy in land plants. UMO-2019/34/E/NZ3/00299
Katarzyna Sieńko, NCN MINIATURA 4 (2021). Odpowiedź transkrypcyjna na indukowany ciemnością proces starzenia się mutantów autofagii Arabidopsis thaliana. K/MNT/000179
Kenji Yamada, FNP TEAM4 (2018-2022). Plant chemical defense based on mustard oil bomb. TEAM/2017-4/41.
Kenji Yamada, NCN OPUS19 (2017-2021). Plant defense system against pathogens and herbivores based on endoplasmic reticulum (ER) body. UMO-2016/23/B/NZ1/01847.
Shino Goto-Yamada, NCN POLONEZ2 (2017-2019). Analysis of quality control of peroxisomes depending on chaperon, protease and autophagy. UMO-2016/21/P/NZ9/01089
Kenji Yamada, NCN OPUS 23 (2023-2026). Molecular and genetic analysis of glucosinolate defence systems and their effect on plant root-microbe interactions. UMO-2022/45/B/NZ3/03700.
Jakub Bizan (technician, 2016-2021)
Toru Maeda (postdoc, 2018-2021)
Shayan Sarkar (postdoc, 2019-2021)
Olga Długosz-Grochowska (postdoc, 2018-2021)
Katarzyna Sieńko (postdoc, 2017-2021)

